Problem class, ground truth annotations and reported metrics
Introduction
To perform benchmarking, ground truth annotations should be encoded in a format that is specific to the associated problem class. BIA workflows are also expected to output results in the same format.
Currently 9 problem classes are supported in BIAFLOWS and their respective annotation formats and computed benchmark metrics are described below.
Note: each problem class has a long name (explicit) and short name (e.g. Object Segmentation / ObjSeg). The same hold for metrics (e.g. DICE / DC).
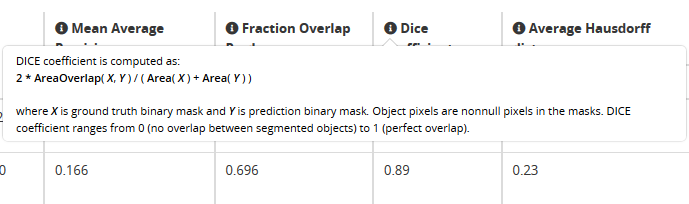
A description of each benchmark is available on the workflow runs result table by clicking on the symbol.
Problem Class
| Problem Class | Tasks | Shortname | Annotation | Example | Metrics | Tools |
|---|---|---|---|---|---|---|
| Object Segmentation | Delineate objects or isolated regions | ObjSeg |
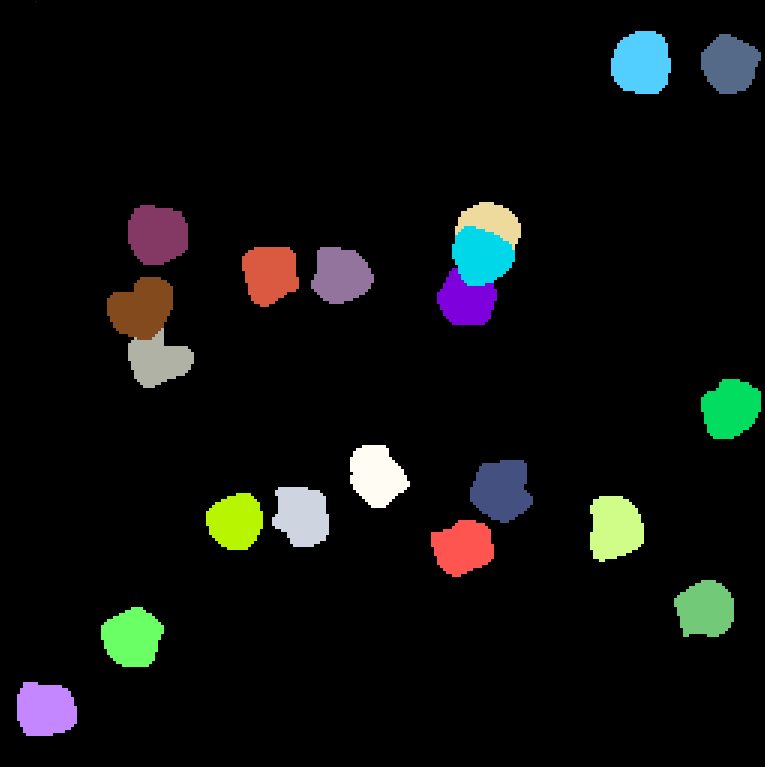 |
sample | ||
| Pixel/Voxel classification | Estimate pixels class | PixCla |
 |
sample | ||
| Spot/Object Counting | Estimate the number of objects | SptCnt |
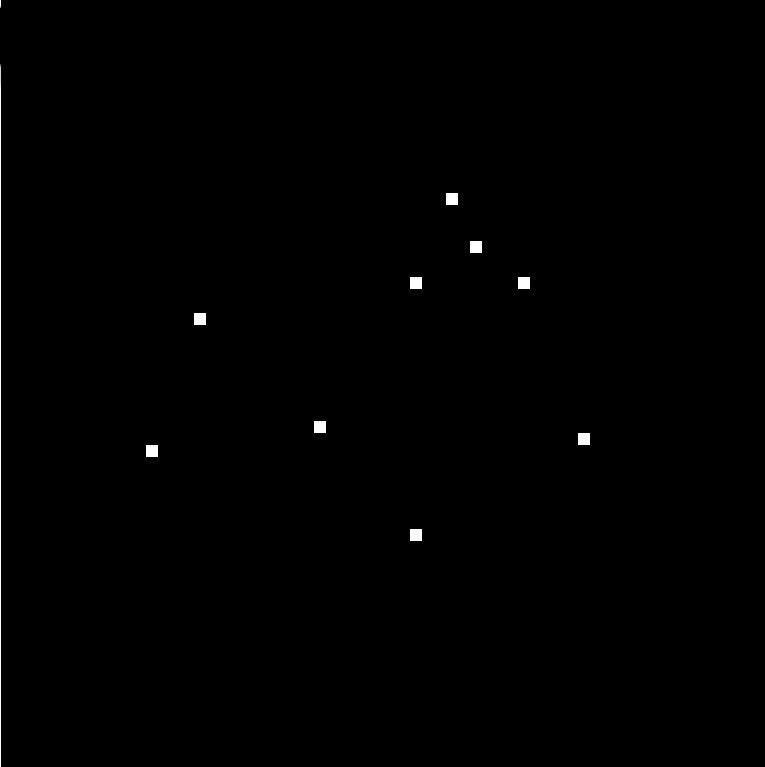 |
sample |
|
|
| Spot/Object Detection | Detect objects in an image (e.g. nucleus) | ObjDet |
 |
sample |
|
|
| Filament Tree Tracing | Estimate the medial axis of a connected filament tree network (one per image) | TreTrc |
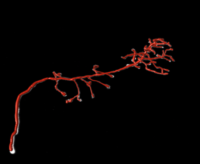 |
sample SWC format |
|
|
| Filament Networks Tracing | Estimate the medial axis of one or several connected filament network(s) | LooTrc |
 |
sample |
|
|
| Landmark Detection | Estimate the position of specific feature points | LndDet |
 |
sample |
|
|
| Particle Tracking | Estimate the tracks followed by particles (no division) | PrtTrk |
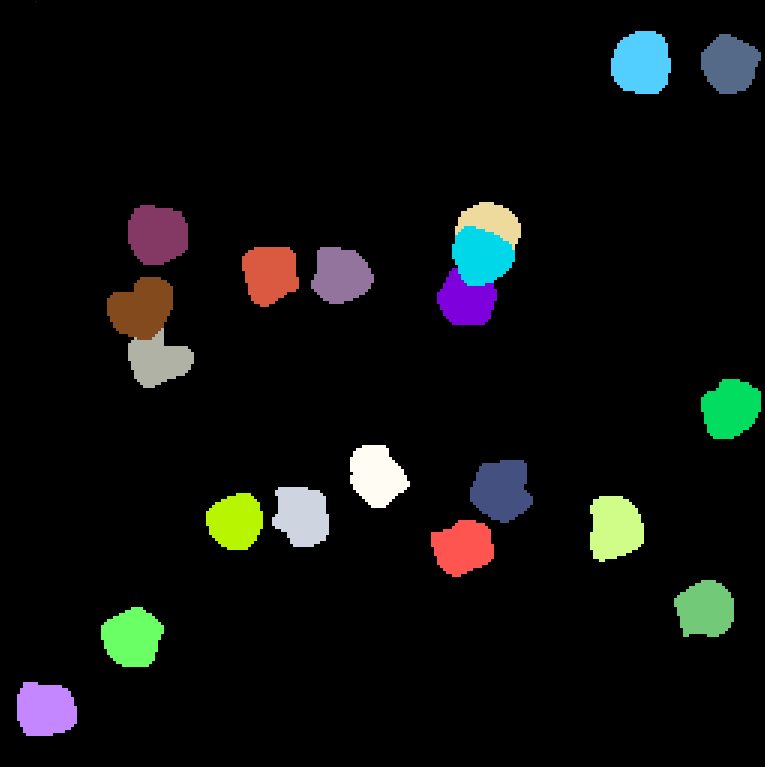 |
sample |
All metric computed by Particle Tracking Challenge Java code (archived here).
|
|
| Object Tracking | Estimate object tracks and segmentation masks (with possible divisions) | ObjTrk |
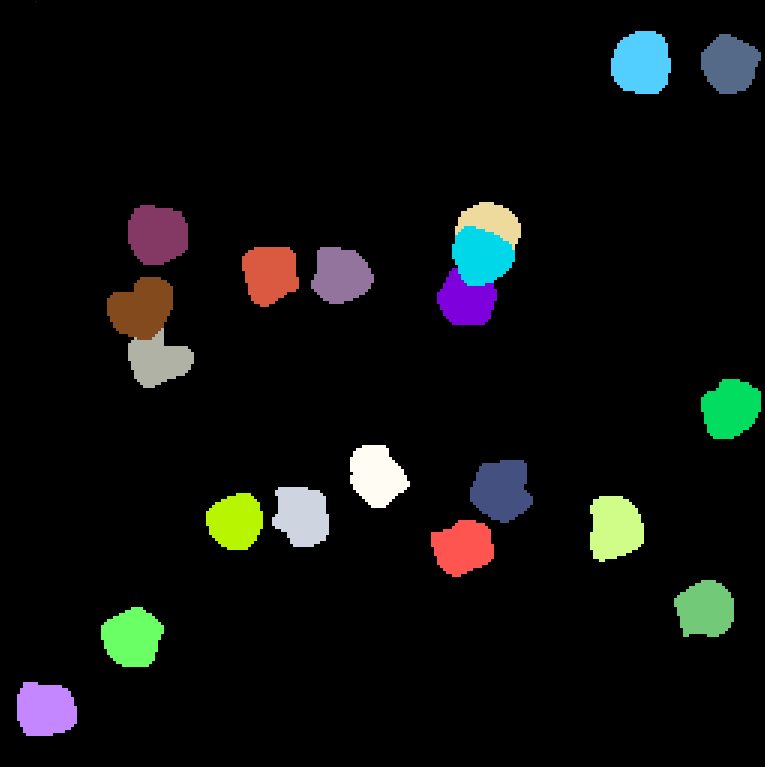 |
sample |
All computed from Cell Tracking Challenge metric command-line executables. |